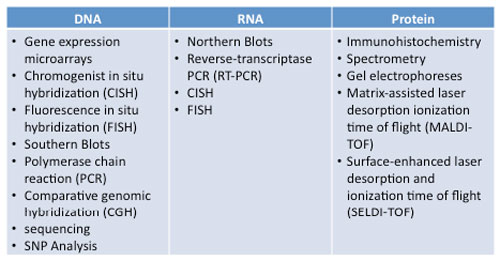
Research Technology - Page 2
Fluorescence In Situ Hybridization (FISH)
FISH is a technique used to detect and localize the presence or absence of specific DNA sequences on chromosomes.
FISH uses fluorescent probes that bind to only those parts of the chromosome with which they show a high degree of sequence similarity. Fluorescence microscopy can be used to find out where the fluorescent probe is bound to the chromosome.
In medicine, FISH can be used to form or confirm a diagnosis, to evaluate a prognosis, or to evaluate remission of a disease, such as cancer. Treatment can then be specifically tailored to the specific situation.
In reseach, FISH can be used to measure chromosome defects like gene deletions and duplications or fusions that can be associated with cancer.
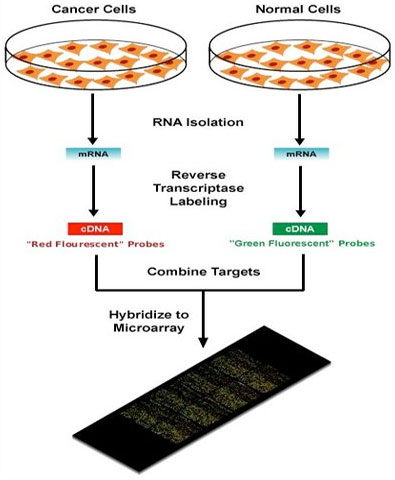
Microarrays
Because this technique is so often discussed in research, this section contains a little more detailed information than the others.
Background:
With only a few exceptions, every cell of the body contains a full set of chromosomes and identical genes. Only a fraction of these genes are turned on or "expressed", however, and it is the subset that is "expressed" that confers unique properties to each cell type.
"Gene expression" is the term used to describe the transcription of the information contained within the DNA, the repository of genetic information, into messenger RNA (mRNA) molecules; these molecules are then transformed into the proteins that perform most of the critical functions of cells.
Scientists study the kinds and amounts of mRNA produced by a cell in order to learn which genes are expressed; this in turn, provides insights into how the cell responds to its changing needs.