Southern blotting
Southern blotting is a method used for probing for the presence of a specific DNA sequence within a DNA sample. Southern blotting is less commonly used in laboratory science due to the ability of other techniques, such as PCR, to detect specific DNA sequences in samples.
These blots are still used for some applications, however, such as measuring copy numbers in transgenic mice, or in the engineering of gene knockout embryonic stem cell lines.
Northern blotting
The northern blot is used to study the expression patterns of a specific type of RNA molecule as relative to others in a set of different samples of RNA.
Southern blot techniques are used to analyze DNA and Northern blot techniques to analyze RNA. In both cases, the nucleic acid is separated by gel electrophoresis, a process that separating molecules on the basis of their size using an electric current applied to a gel matrix.
After gel electrophoresis, the material is transferred to a membrane (blot). The material is then labeled with radio-active probes that bind specifically to the pieces of the nucleic acid that are of interest. Finally, the material marked by radio-active probes is detected by exposure to an x-ray.
This is shown in the picture below.
- Each vertical lane represents a different DNA sample.
- Horizontal bands that end up at the same distance from the top contain molecules that passed through the gel with the same speed, which means they are approximately the same molecular weight.
 |
| Image courtesy of Wikipedia |
DNA sequencing
This technique was developed in 1977 by Frederick Sanger. The term DNA sequencing refers to methods for determining the order of the nucleotide bases adenine, guanine, cytosine, and thymine, in a molecule of DNA.
DNA sequencing has become automated, faster and therefore cheaper. The high demand for low-cost sequencing has driven the development of high-throughput sequencing technologies that simultaneously measures multiple samples (dozens or more) in a single assay (test).
The rapid speed of sequencing attained with modern DNA sequencing technology has been instrumental in allowing the Human Genome Project to sequence the human genome.
In dye-terminator sequencing (image below courtesy of Wikipedia), each of the four DNA bases (T,C,A,G) is labeled with fluorescent dyes. Because each of them have different wavelengths of fluorescence and emission they produce the familiar sequence picture seen in research documents (peaks of different colors with base identified-A,T,G,C).
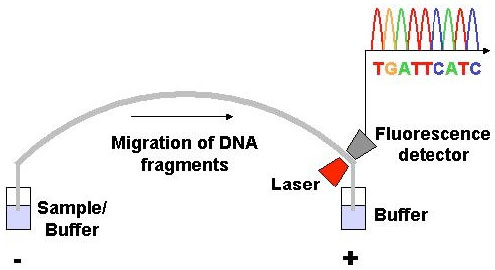 |
| Image courtesy of the National Cancer Institute |
Owing to its greater expediency and speed, dye-terminator sequencing is now the mainstay used in automated sequencing.
Knowledge of the DNA sequences of genes and other elements of the genome of organisms has become indispensable for basic research studying biological processes.
Polymerase chain reaction (PCR)
PCR allows a single DNA sequence to be copied millions of times (amplified), or altered in predetermined ways. There are three basic steps involved in performing a PCR. The steps are repeated 30-40 times in cycles of heating and cooling, with each step taking place at a different temperature. All of the needed components are mixed together in one tube in very tiny volumes. The reaction is carried out in an automated machine, known as a thermocycler, which is capable of rapidly increasing and decreasing the temperature. See image below.
 |
| Image courtesy of the National Cancer Institute |
Applications of PCR
- Allows isolation of DNA fragments from genomic DNA by selective amplification of a specific region of DNA.
- Supplies DNA Sequencing or Southern or Northern Blotting Techniques with high amounts of pure DNA or DNA probes, enabling analysis of DNA samples even from very small amounts of sample material.
- Measures levels or studies the patterns of gene expression
- Researchers can study why something goes wrong in a cell and possibly how to fix it.






