Examples of molecular diagnostics in use today
Non-Hodgkin's lymphoma
Using a chip that contained fragments of 18,000 genes, researchers discovered two
distinct cancer subtypes in a blood cancer called diffuse large B cell lymphoma-the
most common subtype of non-Hodgkin's lymphoma.
These cancers subtypes looked the same under the microscope, but had different
patterns of gene activity. The subtypes were different in other ways, too. For one
thing, they arose from different kinds of cells. Tumor cells of one cancer subtype
arose from less differentiated lymphocytes, while the other subtype arose
from more differentiated lymphocytes.
Later, another gene chip analysis yielded a third and then fourth subtype of diffuse
large B cell lymphoma. Researchers also found 17 genes that were strongly related
to survival.
A formula was created to predict response to chemotherapy based on the expression
patterns of these 17 genes. This formula divided B cell lymphoma patients into four
groups. Two of the groups had about 72% survival rates five years after diagnosis.
The third group had a 34% survival rate, and the fourth had a 15% survival rate.
Breast Cancer
Molecular tests are being offered by several companies, including Genomic Health,
Agendia and AviaraDx to help identify a breast cancer patient's risk of disease
recurrence and/or the likelihood of it becoming metastatic.
These multigene assays are performed on tissue specimens taken from the primary
tumor and utilize molecular methods such as reverse transcriptase polymerase chain
reaction (RT-PCR) to determine gene expression. Depending on the pattern of gene
expression, the patient's risk for recurrence is calculated. In the case of Genomic
Health's Oncotype DX test, the assay is also predictive of response to chemotherapy.
Stomach Cancer
Comparing gene activity patterns in normal stomach tissue and tissue from stomach
tumors, researchers found that the tumor tissue, but not the normal tissue,
expressed a gene called PLA2G2A. They also found that cancer patients with high
expression levels of PLA2G2A were more likely to survive for five years, compared
with patients whose tumors produced lower levels.
Lung Cancer
Using microarrays, researchers discovered that the most common type of lung cancer
called lung adenocarcinoma is actually four distinct types of cancer, each with its
own gene expression pattern.
| |
|
 |
|
| |
Image courtesyof the National Cancer Institute |
|
| |
Colorectal Cancer
Recently, colorectal cancer patients who harbor KRAS mutations were shown to have
no response to antibody-based EGFR therapies such as cetuximab and
panitumumab.
Mutated KRAS genes have been detected in about 40% of metastatic colorectal
cancers. A new clinical laboratory test is now commercially available that identifies
colorectal cancer (CRC) patients who may be resistant to EGFR-targeted monoclonal
antibody therapies.
Reminder: What are tumor markers?
Molecular diagnosis allows for the identification of tumor markers that are used in
the detection, diagnosis, treatment, and monitoring of some types of cancer.
Tumor markers are substances found in the blood, urine, or tissues produced by
tumor cells or other cells in the body in response to cancer or certain benign
conditions. For more information about tumor markers, please go to the CISN fact
sheet called Tumor Markers. (coming soon)
Molecular diagnostics and drug metabolism
Along with cancer diagnosis, molecular diagnostics impact cancer treatment.
Microarrays are being applied to pharmacogenomics-an area of research that
studies why certain drugs work in combination with particular genetic expression
patterns but not with others-to design new drugs that target cancer cells and do not
affect normal ones.
Drugs are broken down in the body by proteins, and not all of these proteins are
identical from person to person. The genetic variations, called SNPs, can result in
subtle differences in proteins that translate to major differences in how the protein
functions.
For example, as many as 40% of people have a SNP that causes a deficiency in a
protein called cytochrome P450 CYP2C9, which is important to drug metabolism.
Deficiencies in this protein can impair the metabolism of as many as 1 in 5
medications currently on the market, such as Tamoxifen, which is used in many ER+
breast cancer patients.
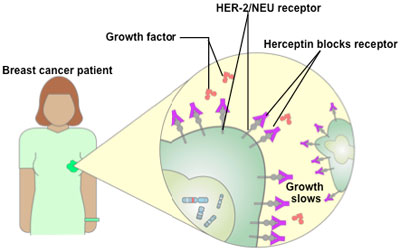
An example of an approved cancer therapy that uses molecular diagnostics
The Food and Drug Administration approved Herceptin in late 1998 as a therapy for
women who test positive for high levels of the Her-2/neu protein, about 25 to 30%
of all breast cancer patients.
Herceptin was developed using pharmacogenomics. In the 1980s, researchers
discovered that some women who had particularly fast-growing breast cancers
expressed extra copies of a gene called Her-2/neu. The genes were producing many copies of a protein that appeared to be driving the growth of the cancer cells.
Some women who were given the experimental antibody saw their cancer growth
slow or stop altogether when their Herceptin treatment was combined with
chemotherapy.